Minibio
Antony Watts

Emeritus Professor of Biochemistry, University of Oxford, UK. Honorary
Research Associate, University of Oxford, (2020 – on). BRIEF RESUMÉ OF BIOPHYSICS RESEARCH ACTIVITY: A highly multidisciplinary biophysical approach has been adopted in the study of proteins and lipids in model and natural membranes. This work has been in three productive phases, beginning with graduate work on thermodynamic studies of lipids in liposomal and model systems (Astbury Department of Biophysics, Leeds, 1972 – 1975), followed by post-doctoral research into structural and functional studies of the molecular specificity of lipid-protein in membranes (Max Plank Institute for Biophysics, Göttingen, Germany, 1976 – 1980). Since returning to an independent faculty position in Oxford (1980), new methodologies have been developed, initially around wide-line NMR (1980-2000) but then high resolution solid state NMR (1994-2009) for the study of biomolecular systems with specific application for resolving high resolution (sub-Å) structural and dynamic details of ligand- and drug-receptor interactions in the absence of other structural information (Watts, 2005, Nature Reviews Drug Discovery, 4, 555-568; invited review).
During this time, most biophysical methods have been employed, and resulting publications include the use of NMR, ESR, ultracentrifugation, diffraction/scattering (X-ray, XFELs, optical and neutron), differential scanning calorimetry, electron microscopy, CD, FRET, ATIR, membrane protein crystallizations, SPR, peptide synthesis and computational approaches, as well as molecular biology for expression, mutagenesis, in-gel fluorescence, PCR, fermentation, directed isotope labelling and novel membrane protein reconstitutions. The general approach is thus to address a system in a “hypothesis driven” approach aimed at understanding biology with a range of appropriate methods, rather than specialize in one method.
68 graduate students have been trained in the laboratory, and over 90 post-docs have been employed at various times – numerous sabbatical workers and collaborators have also been hosted.
Biophysics reveals the importance of water in membrane receptor function – Implications for optogenetics
The physics of living systems has stimulated scientific minds for centuries, but only in the 19th century was it realized that the seemingly chaotic nature of biology, must necessarily obey the exacting laws of physics1. Contemporary biophysics is a highly trans-, cross- and interdisciplinary science, often molecular in nature and with translational impact into areas such as human health, environmental science and synthetic biology.
One contemporary challenge of biophysics is to reveal the intimate molecular and quantum details of how light is converted into energy in biological contexts – nature’s “solar cells”. Light is potentially the most important signal for living organisms, as most of the life on Earth ultimately depends on light energy. Light has been utilized by the earliest of living systems, with early photosynthetic systems emerging ~ 3.5B years ago. One sophistication in light harvesting is the evolution of a family of membrane-embedded proteins called rhodopsins, that developed into sophisticated spectrally-tuned entities around the Great Oxygen Event (GOE), ~ 2.3B years ago2. Fascinating is the very similar structural feature of rhodopsins, with a 7- α helix bundle, but it is the variability in a limited number of amino acid residues that determines wavelength acuity and the functional activity, such as pumping protons, forming selective ion channels, cellular signalling and acting as sensory triggers (as in the eye of higher life forms), all activated though light. Retinal is a common component of rhodopsins, which are the most abundant proteins in the biosphere, and have played a crucial role in the early evolution of life on earth3.
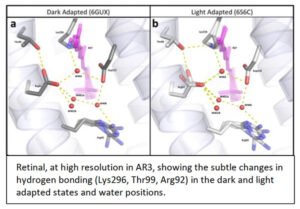 Over the years we have addressed this challenge using a range of spectroscopic approaches4, 5, 6 on functionally competent photoreceptors, often in their natural membranes7 or Lipodisqs™ 8, 9. From functional studies and multiple analytical approaches, we have produced very high resolution (1.07Å) crystallographic structure10, 11, 12 , as well as photo-induced x-ray, free electron laser (XFELS), time-resolved (fs – ms) details, of an achearhodopsin (AR3), without the use of detergents and including natural lipids. This high-resolution information reveals individual waters, as well as their importance in both receptor activation-desensitization, with QM(SCC-DFTB)/MM MD trajectories giving information about the (very fast, fs) activation process following light incidence. AR3 is utilized widely in optogenetics, despite the lack of molecular structures until now. Spectral tuning though mutagenesis, directed from the new structural information, reveals novel wavelength responses, broadening potential utility in optogenetics to more useful wavelengths.
Over the years we have addressed this challenge using a range of spectroscopic approaches4, 5, 6 on functionally competent photoreceptors, often in their natural membranes7 or Lipodisqs™ 8, 9. From functional studies and multiple analytical approaches, we have produced very high resolution (1.07Å) crystallographic structure10, 11, 12 , as well as photo-induced x-ray, free electron laser (XFELS), time-resolved (fs – ms) details, of an achearhodopsin (AR3), without the use of detergents and including natural lipids. This high-resolution information reveals individual waters, as well as their importance in both receptor activation-desensitization, with QM(SCC-DFTB)/MM MD trajectories giving information about the (very fast, fs) activation process following light incidence. AR3 is utilized widely in optogenetics, despite the lack of molecular structures until now. Spectral tuning though mutagenesis, directed from the new structural information, reveals novel wavelength responses, broadening potential utility in optogenetics to more useful wavelengths.
More generally, we suggest that the arrangement of internal water networks in AR3 is responsible for the faster photocycle kinetics compared to homologs – AR3 is ~10x more efficient at current generation than bacteriorhodopsin. These insights may well have generic implications for other receptors including ubiquitous G-protein coupled receptor (GPCRs).
References
- Andersen, O. (2016) Biophys J 110(5) 1028–1032
- Sephus, et al., Mol. Biol. and Evolution (2022), 39;1
- Zabelskii, et al. (2021) Commun Biol 4, 821
- Higman et al., (2011) Angew. Chemie 50(36):8432
- Dijkman et al., (2018) Nature Comms. 9:1710
- Dijkman et al., (2020) Science Advances, 6:33
- Lavington & Watts (2020) Biophys. Rev. 12:1287
- Juarez et al., (2019) Chem. Phys. Lipids 221:167
- Sawczyc et al (2023) Eur. Biophys J. 52:39
- Hoi et al., (2021) Nano Letters, 21(7):2824
- Axford et al., (2022) Acta Cryst D78:52
- Juarez et al (2021) Nature Comms. 12:629

